Lynn Yarris, Lawrence Berkeley Lab
The CRISPR/Cas9 protein system, which is central part to bacterial adaptive immunity, has soared to great prominence in recent years for its enormous potential as a genome editing tool. In studying this system, scientists have found it to be akin to a Russian doll in that the unlocking of one secret reveals another secret within.
Jennifer Doudna, a biochemist with Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging (MBIB) division who has been at the forefront of unlocking CRISPR/Cas secrets, has just unlocked another. Working off data acquired at the Advanced Light Source, Doudna and her research group have discovered the structural basis by which bacteria are able to capture genetic information from viruses and other foreign invaders for use in their own immunological system.
“By studying X-ray crystal structures of Cas1 and Cas2 enzymes in Escherichia coli, we can now see how foreign DNA is manipulated and bent upon being captured by Cas1 and Cas2,” Doudna says. “Knowing how Cas1 and Cas 2 function in bacterial genomes provides us with a possible mechanism for studying or correcting problems in human genomes.”

Credit: Roy Kaltschmidt/Berkeley Lab
Doudna, who also holds appointments with UC Berkeley’s Department of Molecular and Cell Biology and Department of Chemistry, and is an investigator with the Howard Hughes Medical Institute (HHMI), is the corresponding author of a paper in Nature that describes this research. The paper is titled “Foreign DNA capture during CRISPR–Cas adaptive immunity.” James Nuñez and Lucas Harrington, members of Doudna’s research group are the lead authors. Other authors are Philip Kranzusch and Alan Engelman.
While we humans view bacteria as the enemy, bacteria have enemies too – viruses and invading strands of nucleic acid known as plasmids. To protect themselves, bacteria have developed an adaptive-type immune system that revolves around a unit of DNA known as CRISPR, which stands for Clustered Regularly Interspaced Short Palindromic Repeats. A CRISPR unit of DNA is made up of “repeat” elements, base-pair sequences ranging from 30 to 60 nucleotides in length, separated by “spacer” elements, variable sequences that are also from 30 to 60 nucleotides in length. The combination of CRISPR and CRISPR-associated – “Cas” – proteins, enable bacteria to convert spacers into customized RNA molecules that silence critical portions of a foreign invader’s DNA. The CRISPR/Cas system also enables a bacterium to acquire immunity from similar invasions in the future by “remembering” prior infections based on the foreign DNA spacer elements integrated within the bacterium’s CRISPR loci.
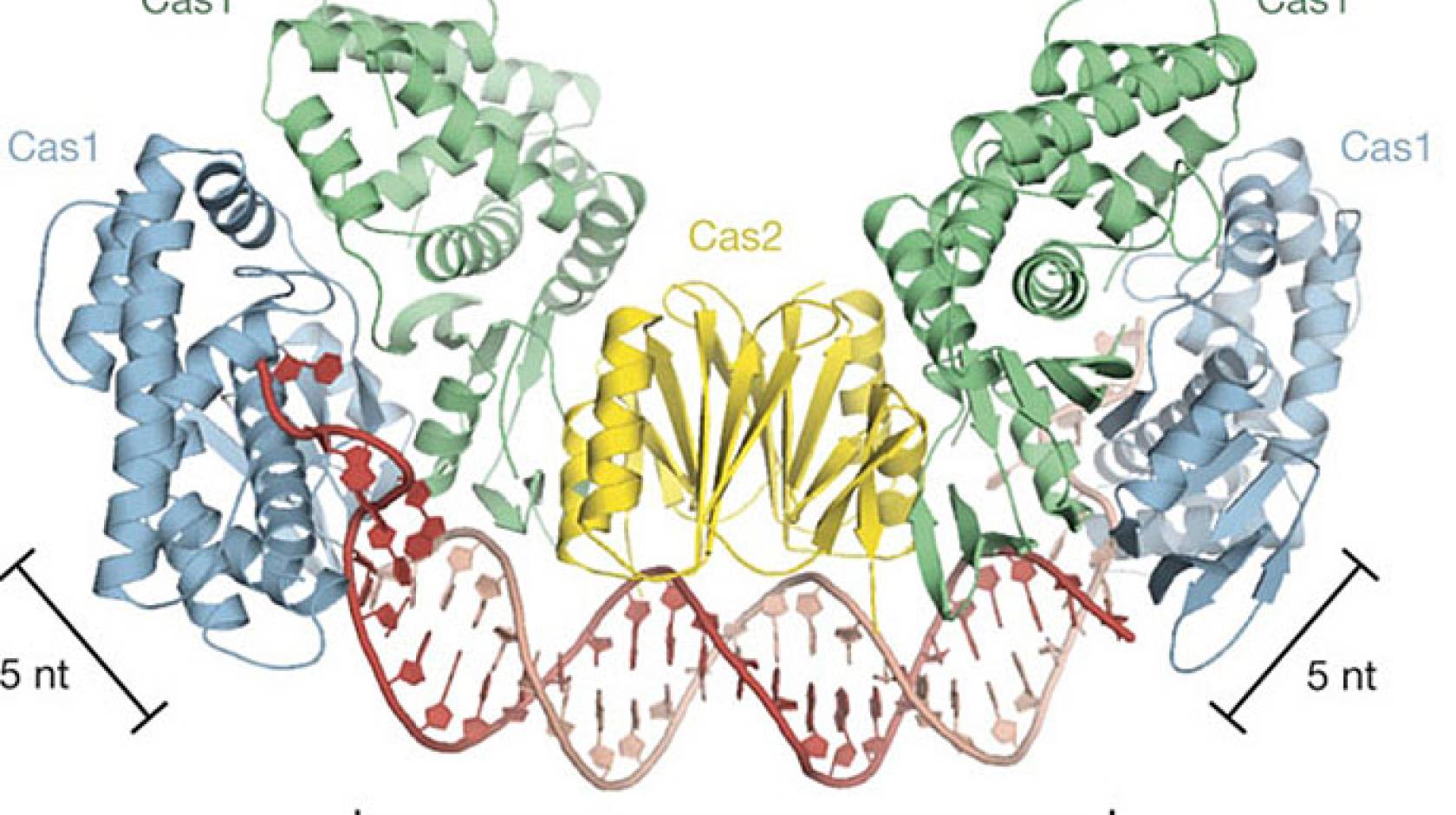
Recently Doudna and her group discovered that Cas1 and Cas2 are the only two proteins in the CRISPR/Cas system required for bacteria to “steal” and “memorize” the genetic information in foreign DNA, but how this task is accomplished remained unknown. Now, using the macromolecular crystallography beamline (8.3.1) at the ALS, which is a U.S. Department of Energy Office of Science user facility, Doudna and her group have discovered that Cas1 and Cas2 function as molecular rulers that will not only recognize foreign DNA but also perfectly measure the DNA during the stealing process.
“We knew from our previous work that Cas1 and Cas2 capture double-stranded DNA instead of single-stranded DNA, but what we didn’t expect when we solved the crystal structure in E. coli was that the ends of the double-stranded DNA are being separated by Cas1,” Nuñez says. “This is a critical finding because we now know we can program Cas1 and Cas2 with DNA substrates containing a central double-stranded DNA region and single-stranded DNA on the ends, and then perhaps insert these DNA substrates into specific sites along a target genome for editing purposes.”
This research was primarily supported by grants from the National Science Foundation.

