Laura Kurtzman, UCSF
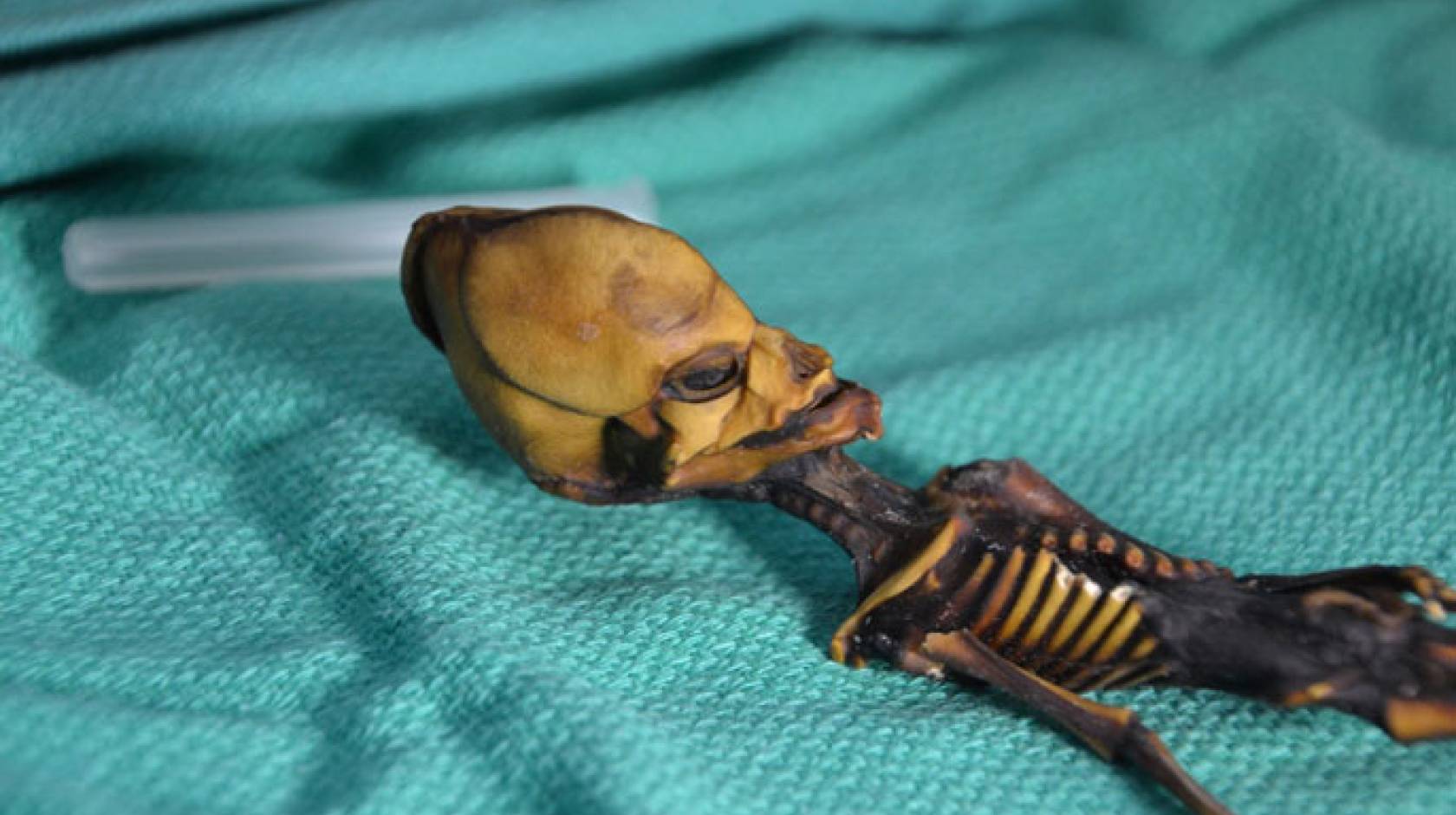
The skeleton, discovered in a leather pouch behind an abandoned church, was pristine: a tiny figure, just six inches long, with a cone-shaped head, 10 pairs of ribs, and bones that looked like those of an eight-year-old child. Found in the Atacama Desert of Chile and later affectionately nicknamed “Ata,” the skeleton made its way onto the black market for archeological finds and then to a collector in Spain who thought it might be the remains of an extraterrestrial being.
But a forensic analysis of Ata’s genome by scientists at UC San Francisco and Stanford University has proved beyond a doubt that it is human. Ata has the DNA of a modern human female with the mix of Native American and European ancestral markers one would expect from someone who lived near the place where she was found. And her arresting appearance, which scientists refer to as a phenotype, can most likely be explained by a handful of rare genetic mutations — some already known, others newly discovered — that are linked to dwarfism and other bone and growth disorders.
Their discovery, published Thursday, March 22, 2018, in Genome Research, does more than lay to rest the fable of Ata’s extraterrestrial origins. It also illustrates how far open-source genetic data has come in enabling the sort of needle-in-a-haystack analysis that can pinpoint the handful of mutated genes — out of more than 2.7 million single-nucleotide variants (SNVs) in Ata’s genome — that were most likely to be associated with the unusual shape of her body.
“The bioinformatics analyses in the paper showcase the power and wealth of information available in the public domain that led to the discovery of novel and rare deleterious variants in genes associated with Ata’s phenotype,” said Sanchita Bhattacharya, a bioinformatics researcher at the UCSF Institute for Computational Health Sciences (ICHS). “The analysis was even more challenging with a very limited amount of information about the specimen, and lack of family history, which makes it a unique case.”
Bhattacharya used the Human Phenotype Ontology (HPO), a database that links genomic data to the abnormal phenotypes found in human disease, everything from atrial septal defect, or a hole in the chambers of the heart, to musculoskeletal abnormalities.
In an initial analysis Bhattacharya found 64 gene variants that seemed likely to be damaging. She fed them into the HPO database, and to her amazement, most of the possible phenotypes the program homed in on had to do with the skeletal system, including “proportionate short stature” and “11 pairs of ribs.” Ata had 10 pairs, a phenotype that had never been observed.
“The moment I saw it, I could see there was something interesting going on there,” Bhattacharya said. “It was a little amount of information, and I am not a bone expert. This was a very blinded analysis.”
New genetic mutations revealed
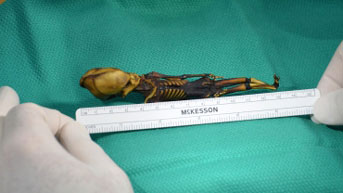
Credit: Emery Smith
The results revealed four new SNVs—a type of genetic mutation at the individual level—in genes that were known to cause bone diseases, like scoliosis or dislocations, as well as two more SNVs in genes involved in producing collagen.
While esoteric, the analysis of Ata’s genome points toward the clinical genetics of the future, said Dr. Atul Butte, Ph.D., who directs ICHS and is the Priscilla Chan and Mark Zuckerberg Distinguished Professor at UCSF.
With rapidly accumulating genetic data, Butte said, scientists can take a “backwards” approach to diagnosis. Instead of starting with a description of the disease and looking for a mutated gene to explain it, they start with the patient’s raw genetic material to see how it differs from a normal, or “reference” set of samples. The genetic variations that pop out of this comparison then reveal, in an unbiased way, what processes are at work in the patient to create disease.
“Analyzing a puzzling sample like the Ata genome can teach us how to handle current medical samples, which may be driven by multiple mutations,” Butte said. “When we study the genomes of patients with unusual syndromes, there may be more than one gene or pathway involved genetically, which is not always considered.”
Garry Nolan, Ph.D., a professor of microbiology and immunology at the Stanford University School of Medicine, began the scientific exploration of Ata in 2012, when a friend called saying he might have found an “alien.”
Nolan believes further research into Ata’s precocious bone aging could one day benefit patients. “Maybe there’s a way to accelerate bone growth in people who need it, people who have bad breaks,” he said. “Nothing like this had been seen before. Certainly, nobody had looked into the genetics of it.”
But Nolan also said that he hopes, one day, little Ata will be given a proper burial. Far from being a visitor from another planet, Ata’s genome marked her as South American, with genetic variations that identified her as being from the Andean region inhabited by the Chilean Chilote Indians. Judging from the skeleton’s intact condition, he said, it is probably no more than 40 years old.
“We now know that it’s a child, and probably either a pre- or post-term birth and death,” he said. “I think it should be returned to the country of origin and buried according to the customs of the local people.”
Other authors of the study include, Matthew Kan and Shann-Ching Chen, of UCSF; Alexandra Sockell, Felice Bava, Xuhuai Ji, Ralph Lachman and Carlos Bustamante, of Stanford; Jian Li, Narges Asadi and Hugo Lam, of Roche Sequencing Solutions; Emery Smith of Ultra Intelligence Corporation; and Maria Avila-Arcos, of the National Autonomous University of Mexico.

